Screening and bioinformatics analysis of key autophagy-related genes in alcoholic hepatitis
-
摘要:
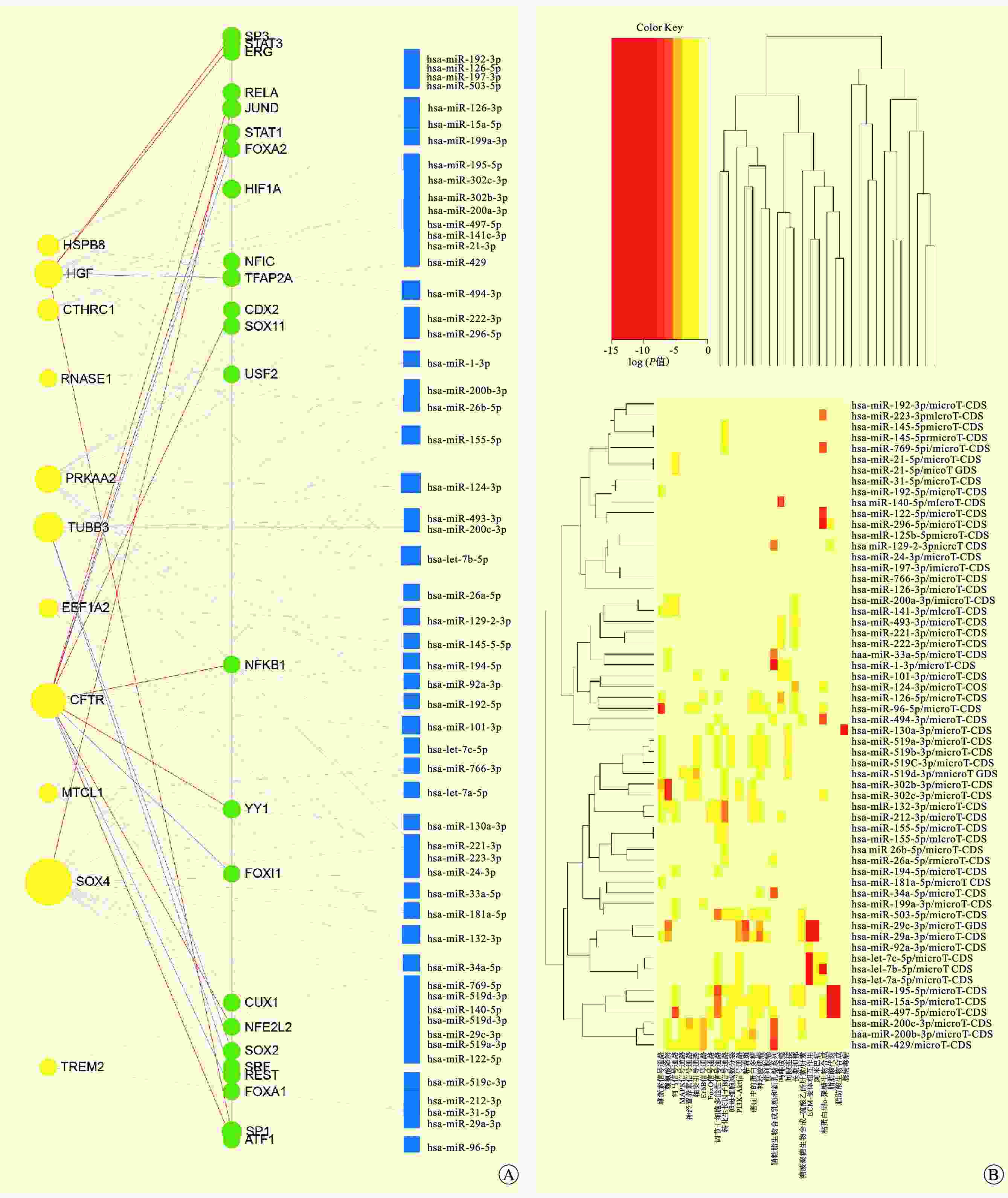
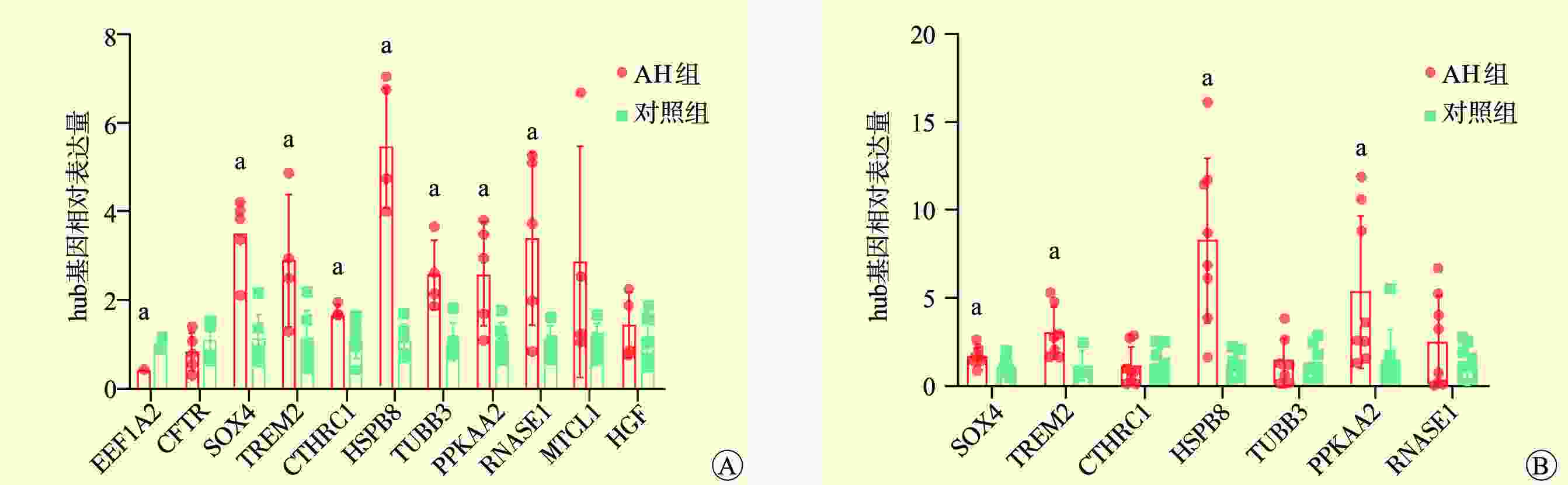
目的 筛选酒精性肝炎(AH)的自噬关键基因,探讨AH潜在的生物标志物和治疗靶点。 方法 采用基因表达综合数据库(GEO)中的2个AH基因芯片和从MSigDB、GeneCards数据库中获得的自噬相关数据集,通过加权基因共表达网络分析(WGCNA)获取关键基因。对筛选的关键基因进行基因本体(GO)、京都基因和基因组百科全书(KEGG)功能富集分析,蛋白质相互作用(PPI)分析,免疫浸润分析,构建信使RNA(mRNA)-微小RNA(miRNA)网络,进行酒精性肝病不同分期的自噬相关关键基因的表达差异分析,并进一步通过实时荧光定量逆转录聚合酶链反应(RT-qPCR)在AH患者和小鼠肝脏组织中验证。 结果 本研究筛选得到了11个与AH自噬相关的基因(EEF1A2、CFTR、SOX4、TREM2、CTHRC1、HSPB8、TUBB3、PRKAA2、RNASE1、MTCL1、HGF),均为上调基因。在AH患者和小鼠肝脏组织中,SOX4、TREM2、HSPB8、PRKAA2在AH组中的相对表达量均高于对照组。 结论 SOX4、TREM2、HSPB8、PRKAA2可能是AH潜在的生物标志物和治疗靶点。 -
关键词:
- 酒精性肝炎 /
- 自噬 /
- 关键基因 /
- 生物信息学 /
- 加权基因共表达网络分析(WGCNA) /
- 基因本体(GO) /
- 京都基因和基因组百科全书(KEGG) /
- 蛋白质相互作用(PPI)
Abstract:Objective To screen key autophagy-related genes in alcoholic hepatitis (AH) and investigate potential biomarkers and therapeutic targets for AH. Methods Two AH gene chips in Gene Expression Omnibus (GEO) and autophagy-related data sets obtained from MSigDB and GeneCards databases were used, and the key genes were verified and obtained by weighted gene co-expression network analysis (WGCNA). The screened key genes were subject to gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), protein-protein interaction (PPI) and immune infiltration analyses. Messenger RNA (mRNA)- microRNA (miRNA) network was constructed to analyze the expression differences of key autophagy-related genes during different stages of AH, which were further validated by real-time fluorescence quantitative polymerase chain reaction (RT-qPCR) in the liver tissues of AH patients and mice. Results Eleven autophagy-related genes were screened in AH (EEF1A2, CFTR, SOX4, TREM2, CTHRC1, HSPB8, TUBB3, PRKAA2, RNASE1, MTCL1 and HGF), all of which were up-regulated. In the liver tissues of AH patients and mice, the relative expression levels of SOX4, TREM2, HSPB8 and PRKAA2 in the AH group were higher than those in the control group. Conclusions SOX4, TREM2, HSPB8 and PRKAA2 may be potential biomarkers and therapeutic targets for AH. -
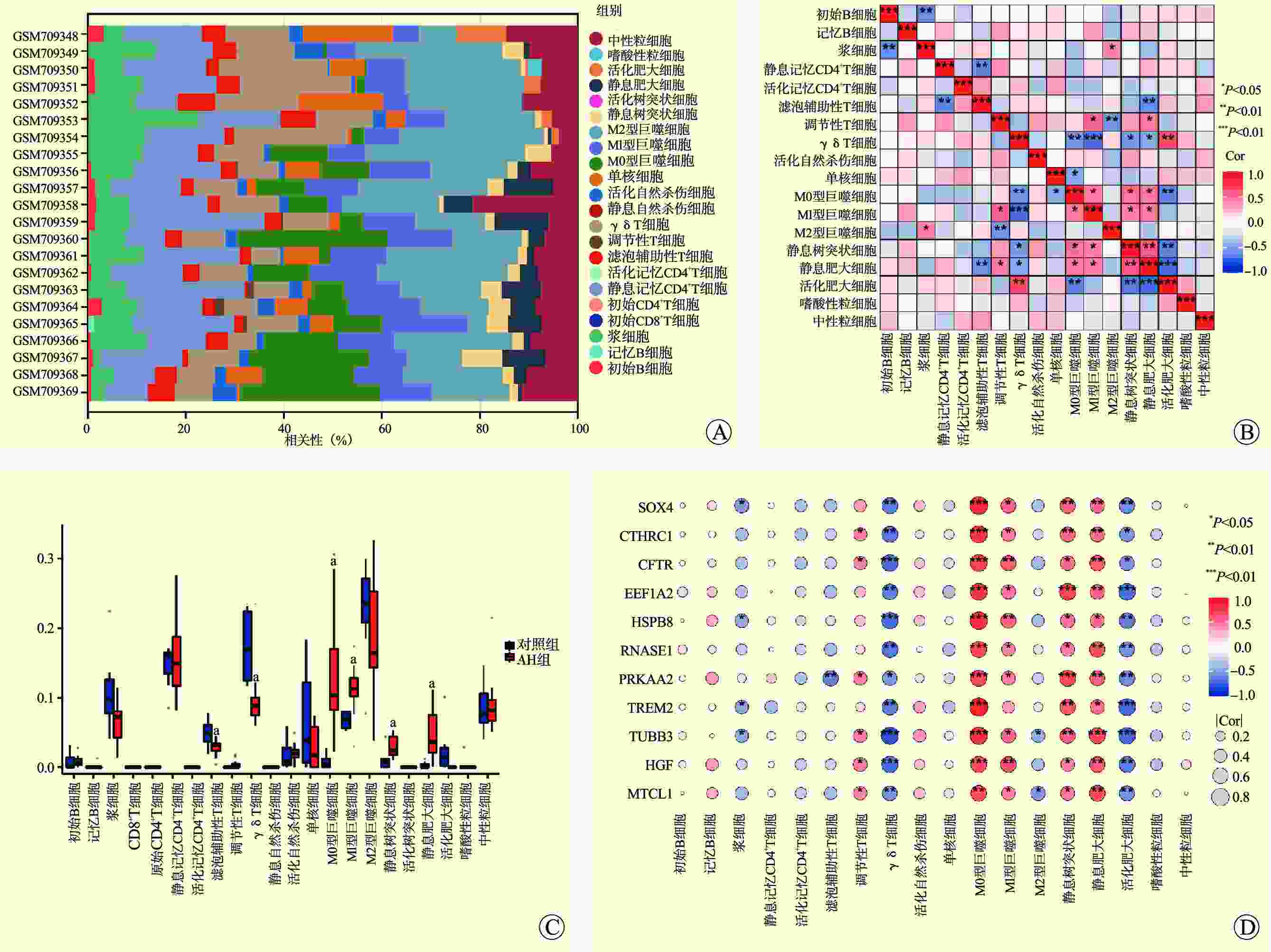
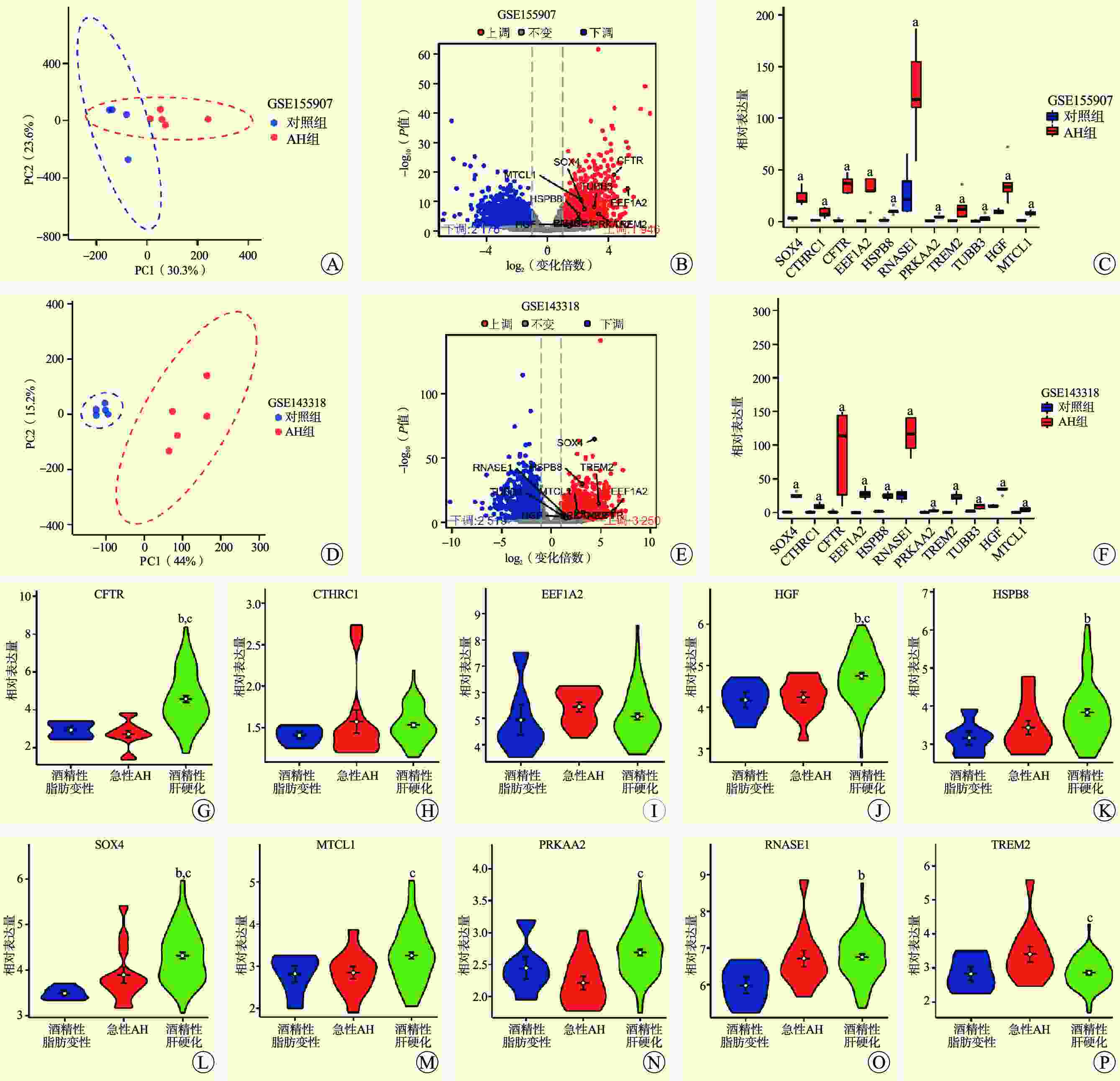
图 5 GSE155907、GSE143318数据集验证和AH不同分期的基因表达
注:A图为GSE155907数据集PCA的结果图;B图示GSE155907数据集火山图验证了11个基因均上调;C图为GSE155907数据集中11个基因表达情况箱形图;D图为GSE143318数据集PCA的结果图;E图示GSE143318数据集火山图验证了11个基因均上调;F图为GSE143318数据集中11个基因表达情况箱形图;G~P图示酒精性肝病不同分期的10种hub基因表达;与对照组比较,aP<0.05;与酒精性脂肪变性比较,bP<0.05;与急性AH比较,cP<0.05。
Figure 5. Datasets validation of GSE155907 and GSE143318 and gene expression of AH at different stages
-
[1] 刘露露, 佟静, 王炳元. 酒精性肝炎临床治疗进展[J]. 中华肝脏病杂志, 2022, 30(6): 672-675. DOI: 10.3760/cma.j.cn501113-20210102-00003.LIU LL, TONG J, WANG BY. Progress in the clinical treatment of alcoholic hepatitis[J]. Chin J Hepatol, 2022, 30(6): 672-675. DOI: 10.3760/cma.j.cn501113-20210102-00003. [2] SEITZ HK, STICKEL F. Molecular mechanisms of alcohol-mediated carcinogenesis[J]. Nat Rev Cancer, 2007, 7(8): 599-612. DOI: 10.1038/nrc2191. [3] DING WX, LI M, CHEN X, et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice[J]. Gastroenterology, 2010, 139(5): 1740-1752. DOI: 10.1053/j.gastro.2010.07.041. [4] PUROHIT V, GAO B, SONG BJ. Molecular mechanisms of alcoholic fatty liver[J]. Alcohol Clin Exp Res, 2009, 33(2): 191-205. DOI: 10.1111/j.1530-0277.2008.00827.x. [5] PONE EJ, ZAN H, ZHANG J, et al. Toll-like receptors and B-cell receptors synergize to induce immunoglobulin class-switch DNA recombination: relevance to microbial antibody responses[J]. Crit Rev Immunol, 2010, 30(1): 1-29. DOI: 10.1615/critrevimmunol.v30.i1.10. [6] ASRANI SK, DEVARBHAVI H, EATON J, et al. Burden of liver diseases in the world[J]. J Hepatol, 2019, 70(1): 151-171. DOI: 10.1016/j.jhep.2018.09.014. [7] LEFKOWITCH JH. Morphology of alcoholic liver disease[J]. Clin Liver Dis, 2005, 9(1): 37-53. DOI: 10.1016/j.cld.2004.11.001. [8] TELI MR, DAY CP, BURT AD, et al. Determinants of progression to cirrhosis or fibrosis in pure alcoholic fatty liver[J]. Lancet, 1995, 346(8981): 987-990. DOI: 10.1016/s0140-6736(95)91685-7. [9] SINGAL AK, MATHURIN P. Diagnosis and treatment of alcohol-associated liver disease: a review[J]. JAMA, 2021, 326(2): 165-176. DOI: 10.1001/jama.2021.7683. [10] 李玉贤, 任锋, 陈煜. 细胞自噬对肝脏疾病代谢的调节作用研究进展[J]. 中华肝脏病杂志, 2023, 31(1): 105-108. DOI: 10.3760/cma.j.cn501113-20201106-00601.LI YX, REN F, CHEN Y. Research progress of the regulatory role of autophagy in metabolic liver diseases[J]. Chin J Hepatol, 31(1): 105-108. DOI: 10.3760/cma.j.cn501113-20201106-00601. [11] MATHURIN P. Early liver transplantation for acute alcoholic hepatitis: we can't say no[J]. J Hepatol, 2021, 75(3): 718-722. DOI: 10.1016/j.jhep.2021.05.019. [12] YANG C, MEI H, PENG L, et al. Prognostic correlation of an autophagy-related gene signature in patients with head and neck squamous cell carcinoma[J]. Comput Math Methods Med, 2020: 7397132. DOI: 10.1155/2020/7397132. [13] VARGAS JNS, HAMASAKI M, KAWABATA T, et al. The mechanisms and roles of selective autophagy in mammals[J]. Nat Rev Mol Cell Biol, 2023, 24(3): 167-185. DOI: 10.1038/s41580-022-00542-2. [14] 代倩兰, 刘绍能. 细胞自噬及其在肝纤维化中的作用[J]. 临床肝胆病杂志, 2021, 37(6): 1440-1444. DOI: 10.3969/j.issn.1001-5256.2021.06.046.DAI QL, LIU SN. Role of autophagy in liver fibrosis[J]. J Clin Hepatol, 2021, 37(6): 1440-1444. DOI: 10.3969/j.issn.1001-5256.2021.06.046. [15] LI W, HE P, HUANG Y, et al. Selective autophagy of intracellular organelles: recent research advances[J]. Theranostics, 2021, 11(1): 222-256. DOI: 10.7150/thno.49860. [16] KOTANI T, SAKAI Y, KIRISAKO H, et al. A mechanism that ensures non-selective cytoplasm degradation by autophagy[J]. Nat Commun, 2023, 14(1): 5815. DOI: 10.1038/s41467-023-41525-x. [17] LUCANTONI F, MARTÍNEZ-CEREZUELA A, GRUEVSKA A, et al. Understanding the implication of autophagy in the activation of hepatic stellate cells in liver fibrosis: are we there yet?[J]. J Pathol, 2021, 254(3): 216-228. DOI: 10.1002/path.5678. [18] MARTINEZ-LOPEZ N, SINGH R. Autophagy and lipid droplets in the liver[J]. Annu Rev Nutr, 2015, 35: 215-237. DOI: 10.1146/annurev-nutr-071813-105336. [19] KIM J, LEE Y, JEON T, et al. All cells are created equal in the sight of autophagy: selective autophagy maintains homeostasis in senescent cells[J]. Autophagy, 2021, 17(10): 3260-3261. DOI: 10.1080/15548627.2021.1953848. [20] SALETE-GRANADO D, CARBONELL C, PUERTAS-MIRANDA D, et al. Autophagy, oxidative stress, and alcoholic liver disease: a systematic review and potential clinical applications[J]. Antioxidants (Basel), 2023, 12(7): 1425. DOI: 10.3390/antiox12071425. [21] ZHAO C, FU Y, MA X, et al. Autophagy-based intervention strategy in the management of hepatotoxicity[J]. Antioxid Redox Signal, 2023, 38(16/18): 1082-1100. DOI: 10.1089/ars.2022.0051. [22] HOSSEINI N, SHOR J, SZABO G. Alcoholic hepatitis: a review[J]. Alcohol Alcohol, 2019, 54(4): 408-416. DOI: 10.1093/alcalc/agz036. [23] THANDA HAN MA, PYRSOPOULOS N. Emerging therapies for alcoholic hepatitis[J]. Clin Liver Dis, 2021, 25(3): 603-624. DOI: 10.1016/j.cld.2021.03.006. [24] ZHOU JC, WANG JL, REN HZ, et al. Autophagy plays a double-edged sword role in liver diseases[J]. J Physiol Biochem, 2022, 78(1): 9-17. DOI: 10.1007/s13105-021-00844-7. [25] QIAN H, CHAO X, WILLIAMS J, et al. Autophagy in liver diseases: a review[J]. Mol Aspects Med, 2021, 82: 100973. DOI: 10.1016/j.mam.2021.100973. [26] 程金来, 周子玉, 刘丽,等. 小鼠急性酒精性肝损伤造模方法的比较[J]. 中国比较医学杂志, 2023, 33(7): 26-33,40. DOI: 10.3969/j.issn.1671-7856.2023.07.004.CHENG JL, ZHOU ZY, LIU L, et al. Comparison and optimization of acute alcoholic liver injury model in mice[J]. Chin J Comp Med, 2023, 33(7): 26-33,40. DOI: 10.3969/j.issn.1671-7856.2023.07.004. [27] ZENG T, ZHANG CL, SONG FY, et al. PI3K/Akt pathway activation was involved in acute ethanol-induced fatty liver in mice[J]. Toxicology, 2012, 296(1/3): 56-66. DOI: 10.1016/j.tox.2012.03.005. [28] BALLESTEROS-ÁLVAREZ J, ANDERSEN JK. mTORC2: the other mTOR in autophagy regulation[J]. Aging Cell, 2021, 20(8): e13431. DOI: 10.1111/acel.13431. [29] SINHA RA. Autophagy: a cellular guardian against hepatic lipotoxicity[J]. Genes (Basel), 2023, 14(3): 553. DOI: 10.3390/genes14030553. [30] CHENG Z. The FoxO-autophagy axis in health and disease[J]. Trends Endocrinol Metab, 2019, 30(9): 658-671. DOI: 10.1016/j.tem.2019.07.009. [31] GUO X, MA X, XUE L. A conserved interplay between FOXO and SNAI/snail in autophagy[J]. Autophagy, 2022, 18(11): 2759-2760. DOI: 10.1080/15548627.2022.2063559. [32] HUANG J, KANG S, PARK SJ, et al. Apelin protects against liver X receptor-mediated steatosis through AMPK and PPARα in human and mouse hepatocytes[J]. Cell Signal, 2017, 39: 84-94. DOI: 10.1016/j.cellsig.2017.08.003. [33] GENG J, BABA M, NAIR U, et al. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy[J]. J Cell Biol, 2008, 182(1): 129-140. DOI: 10.1083/jcb.200711112. [34] FaGHFOURI AH, KHAJEBISHAK Y, PAYAHOO L, et al. PPAR-gamma agonists: potential modulators of autophagy in obesity[J]. Eur J Pharmacol, 2021, 912: 174562. DOI: 10.1016/j.ejphar.2021.174562. [35] CHEN X, MAO R, SU W, et al. Circular RNA circHIPK3 modulates autophagy via miR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer[J]. Autophagy, 2020, 16(4): 659-671. DOI: 10.1080/15548627.2019.1634945. [36] JIAO Y, ZHAO J, ZHANG Z, et al. SRY-box containing gene 4 promotes liver steatosis by upregulation of SREBP-1c[J]. Diabetes, 2018, 67(11): 2227-2238. DOI: 10.2337/db18-0184. [37] LI XC, HU QK, CHEN L, et al. HSPB8 promotes the fusion of autophagosome and lysosome during autophagy in diabetic neurons[J]. Int J Med Sci, 2017, 14(13): 1335-1341. DOI: 10.7150/ijms.20653. [38] LU X, XUAN W, LI J, et al. AMPK protects against alcohol-induced liver injury through UQCRC2 to up-regulate mitophagy[J]. Autophagy, 2021, 17(11): 3622-3643. DOI: 10.1080/15548627.2021.1886829. [39] XU YL, LIU XY, CHENG SB, et al. Geniposide enhances macrophage autophagy through downregulation of TREM2 in atherosclerosis[J]. Am J Chin Med, 2020, 48(8): 1821-1840. DOI: 10.1142/S0192415X20500913. [40] LI RY, QIN Q, YANG HC, et al. TREM2 in the pathogenesis of AD: a lipid metabolism regulator and potential metabolic therapeutic target[J]. Mol Neurodegener, 2022, 17(1): 40. DOI: 10.1186/s13024-022-00542-y. -
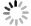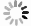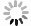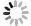




 下载:
下载: